2162 molecular structures approved between 1939 and 2025 with a molecular weight ≤ 2000 have been registered (last update: August 2025).
References:
- Data sets representative of the Structures and Experimental Properties of FDA-approved Drugs, Douguet D., ACS Med Chem Lett., 2018, 9(3):204-209. doi: 10.1021/acsmedchemlett.7b00462
- Pihan E., Colliandre L., Guichou J.-F. and Douguet D., e-Drug3D: 3D structure collections dedicated to drug repurposing and fragment-based drug design, Bioinformatics, 2012, 28(11):1540-1541. doi: 10.1093/bioinformatics/bts186
- Former versions of data sets are available on Zenodo
August, 27, 2025
 DORDAVIPRONE
DORDAVIPRONE
- Browse the list of registered drug structures
- Browse the database (the last 2 registered drugs are displayed)
- Browse / Search the database by text or by chemical substructure (and create subsets to download)
- Browse the WHO ATC tree of registred drugs
- Browse drug names ranked in descending order of approval year or in alphabetical order
- Zoom on the Approved peptides
- Statistics
- Version history
- Chemical Structures (component 1/4):
- Browse / Search the database by text or by chemical substructure
- Download the current version of the e-Drug3D collection (sdf format file) ; one 3D conformer ; ionization of carboxylic acid, phosphate, phosphonate, phosphonoamide, amidinium and guanidinium groups. The datablock contains the ID, name (INN), CAS number and Status.
You can install and use DataWarrior (free software) to read the file content as a table (available for Windows/Linux/MacOS-X)
- Virtually screen the drug structure data set (Docking, properties and shape similarity functions are available)
- Browse / Search the database by text or by chemical substructure
- Pharmacodynamics (component 2/4)
- Download the e-Drug3D-PD data set in text format. Column/field value is separated by a semicolon. It contains the e-Drug3D ID, INN (drug name), CAS number, year of approval, Status, Primary target, ATC code(s), PDB code and ligand code when cocrystallized and list of targets from DrugCentral
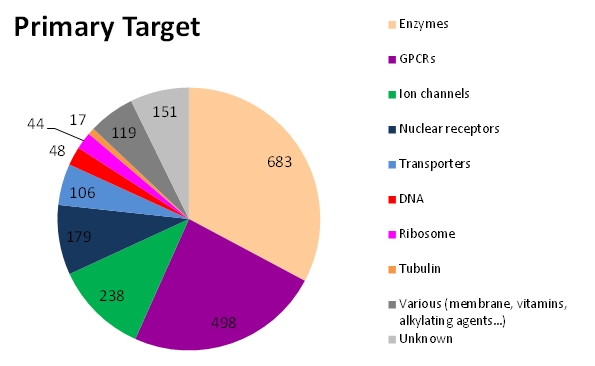
- Download the e-Drug3D-PD data set in text format. Column/field value is separated by a semicolon. It contains the e-Drug3D ID, INN (drug name), CAS number, year of approval, Status, Primary target, ATC code(s), PDB code and ligand code when cocrystallized and list of targets from DrugCentral
- Pharmacokinetics (component 3/4)
- Display metabolite groups (drugs and active metabolites belonging to the same family have been gathered in the same group. Thus, a structure is either a mother or an offspring of another one)
- Download the e-Drug3D-PK data set in text format. Column/field value is separated by a semicolon. It contains the e-Drug3D ID, INN (drug name), CAS number, year of approval, Status, is_or_has a metabolite, routes of administration, Volume of distribution (VD), Clearance (Cl), Plasma Protein Binding (PPB), Half-life (t1/2), Bioavailability (F), Cmax/Tmax, comment on solubility.
- Display metabolite groups (drugs and active metabolites belonging to the same family have been gathered in the same group. Thus, a structure is either a mother or an offspring of another one)
- FDA Registration Data (component 4/4)
- Download the e-Drug3D-RD data set in text format. Column/field value is separated by a semicolon. It contains the ID, name (INN), CAS number, First year of approval, Status, KNApSAcK or NP-Atlas Id if natural product, all associated NDA numbers [FDA approval number, name of the label file in PDF format, company name, year of approval and commercial name of the drug] and the Indication/Therapeutic class information
- Download the drug label files in PDF format (compressed directory). A label file is named with the NDA number. The NDA number is the approval number assigned by the FDA. A drug may possess several NDA numbers (see the above e-Drug3D-RD data set)
- The following file contains the list of discarded NDA numbers as of August 2025 (biologics, contrast agents...)
The current content of the database in terms of number of:
| Properties extracted from the drug's labels | Number of values | ||
| Entries | Structures | enantiomers have different structures | 2162 |
| Princeps | New Molecular Entity (NME) | (i.e. different names) | 1712 |
| is a metabolite | active metabolite | 296 a | |
| has a metabolite | gives an active metabolite | 315 a | |
| Route | Route of administration | 1964 b | |
| PK (Human) | VD | volume of distribution (liter) | 1123 |
| Cl | clearance (liter/hour) | 1123 | |
| t1/2 | half-life (hour) | 1533 | |
| PPB | plasma protein binding (%) | 1326 | |
| F | bioavailability (%) | 603 | |
| Cmax | maximal concentration in blood | 976 | |
| Cmax unit | Cmax unit | 976 | |
| Tmax | time to reach the Cmax (hour) | 975 | |
| Solubility | Comment on experimental solubility | 1134 | |
| PD | Target | Primary target(s) or mechanism of action | 1934 |
| PDB coordinates | Co-structure of the drug with a protein | 845 | |
| a 27% of structures are or have active metabolite(s) - 17 drugs are in both categories b The 198 absent structures are active metabolites without NDA numbers | |||
Drug names (INN) are listed below:
They have been gathered in function of the year of the first approval in decreasing order.
The name of the pharmaceutical company is given in parentheses.
Dxxxx is the identifier of the drug in our database.
'M' in front of a name means that the compound is (an active) METABOLITE (at bottom of the list).
The year 0 lists drugs without any defined year (mainly withdrawn or discontinued drugs but with valid NDA agreement number)
|
Browse Drug names (INN) in function of the year of the first approval: list.html
|
Browse Drug names (INN) in alphabetical order: list.html
|
Description of the database:
- e-Drug3D mirrors the current content of the U.S. pharmacopeia of small drugs (molecular weight ≤ 2000). Discarded NDA/Approved Drugs (biologics, contrast agents...) are listed at this link.
- e-Drug3D uses the 'Drugs@FDA Data File' (http://www.fda.gov/Drugs/InformationOnDrugs/ucm079750.htm) released by the FDA to construct and update the database.
- The structure of each drug is manually drawn and checked to assign the exact stereochemistry to chiral centers (enantiomers are different structures and are registered as such). Chemical structures have been extracted from drug labels and checked using the SciFinder/CAS database.
- Active metabolites have been registered with the prefix "M " (except if the active metabolite structure has been approved under another name or possesses an INN).
- FDA registration data are provided with a link to the FDA website.
- Pharmacokinetic data has been manually extracted from the drug label (additional sources were the review by Obach R.S. et al. (Drug Metabolism and Disposition, 2008, 36(7), 1385-1405. doi: 10.1124/dmd.108.020479 ), PubMed and the EMA).
- Pharmacodynamic data (Primary target) has been manually extracted from the drug label (additional sources are PubMed and DrugCentral).
- A link to the Anatomical Therapeutic Chemical (ATC) classification system (World Health Organization) is provided when it exists.
- Physicochemical properties have been calculated.
- Chemistry information and links to patents, synthesis references and commercial suppliers are indicated.

