| | ||||||||||
| 1 | INN | Class | Route (list) | PK parameters= Cmax; Tmax; F: bioavailability; t1/2: half-life; VD: volume of distribution; Cl: clearance; PPB: plasma protein binding;(EQN means that the equation t1/2 = VD / Cl * 0.693 was used | Primary Target and PDB code of Protein-Drug complex | Targets from DrugCentral | Links | |||
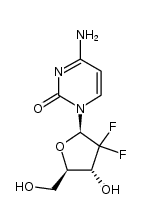
| GEMCITABINE (has an active metabolite)
| GEMCITABINE | ANTINEOPLASTIC | INJECTION INTRAVENOUS | VD 80 LITER PPB 0 PERCENT Cl 88 LITER / HOUR HT 1 HOUR SOLUBILITY SOLUBLE IN WATER | DEOXYCYTIDINE KINASE PDB 2NO0 (C4S DCK VARIANT OF DCK IN COMPLEX WITH GEMCITABINE+ADP) LIGAND CODE = GEO (link to the list of PDB complexes) Download experimental 3D coordinates of GEO with added hydrogens | Ribonucleoside-diphosphate reductase large subunit UNIPROT P23921 RRM1 -- Ribonucleoside-diphosphate reductase subunit M2 UNIPROT P31350 RRM2 more at DrugCentral | EMA | |||
| 2 | INN | Class | Route (list) | PK parameters= Cmax; Tmax; F: bioavailability; t1/2: half-life; VD: volume of distribution; Cl: clearance; PPB: plasma protein binding;(EQN means that the equation t1/2 = VD / Cl * 0.693 was used | Primary Target and PDB code of Protein-Drug complex | Targets from DrugCentral | Links | |||
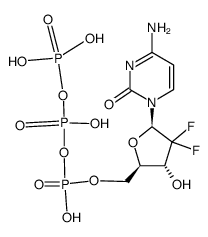
| M GEMCITABINE (is an active metabolite)
| GEMCITABINE | ANTINEOPLASTIC | - | DEOXYCYTIDINE KINASE PDB 2EUD (STRUCTURES OF YEAST RIBONUCLEOTIDE REDUCTASE I COMPLEXED WITH LIGANDS AND SUBUNIT PEPTIDES) LIGAND CODE = GCQ (link to the list of PDB complexes) Download experimental 3D coordinates of GCQ with added hydrogens | Ribonucleoside-diphosphate reductase large subunit UNIPROT P23921 RRM1 -- Ribonucleoside-diphosphate reductase subunit M2 UNIPROT P31350 RRM2 more at DrugCentral | EMA | ||||
| 3 | INN | Class | Route (list) | PK parameters= Cmax; Tmax; F: bioavailability; t1/2: half-life; VD: volume of distribution; Cl: clearance; PPB: plasma protein binding;(EQN means that the equation t1/2 = VD / Cl * 0.693 was used | Primary Target and PDB code of Protein-Drug complex | Targets from DrugCentral | Links | |||
| M GEMCITABINE (is an active metabolite)
| GEMCITABINE | ANTINEOPLASTIC | - | DEOXYCYTIDINE KINASE PDB 3MDC (DNA POLYMERASE LAMBDA IN COMPLEX WITH DFDCTP) LIGAND CODE = GTF (link to the list of PDB complexes) Download experimental 3D coordinates of GTF with added hydrogens | Ribonucleoside-diphosphate reductase large subunit UNIPROT P23921 RRM1 -- Ribonucleoside-diphosphate reductase subunit M2 UNIPROT P31350 RRM2 more at DrugCentral | EMA | ||||