| | ||||||||||
| 1 | INN | Class | Route (list) | PK parameters= Cmax; Tmax; F: bioavailability; t1/2: half-life; VD: volume of distribution; Cl: clearance; PPB: plasma protein binding;(EQN means that the equation t1/2 = VD / Cl * 0.693 was used | Primary Target and PDB code of Protein-Drug complex | Targets from DrugCentral | Links | |||
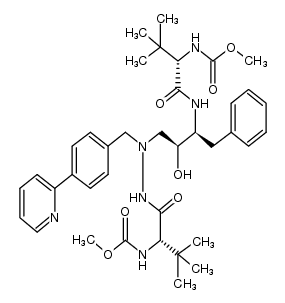
| ATAZANAVIR
| ATAZANAVIR |
ANTIINFECTIVES | ORAL | Cmax 8.7 MICROMOLAR Tmax 2.5 HOUR PPB 86 PERCENT HT 7.9 HOUR SOLUBILITY SLIGHTLY SOLUBLE IN WATER 4 TO 5 MILLIGRAM / MILLILITER WITH THE PH=1.9 AT 24 DEGREE CELSIUS | HIV-1 PROTEASE PDB 3EKW (CRYSTAL STRUCTURE OF THE INHIBITOR ATAZANAVIR (ATV) IN COMPLEX WITH A MULTI-DRUG RESISTANCE HIV-1 PROTEASE VARIANT (L10I, G48V, I54V, V64I, V82A) REFER: FLAP+ IN CITATION) LIGAND CODE = DR7 (link to the list of PDB complexes) Download experimental 3D coordinates of DR7 with added hydrogens | Pol polyprotein UNIPROT Q72874 pol more at DrugCentral | EMA | |||