Download Molecule MRTX1133: 7t47_B_6IC.sdf extracted from the PDB complex 7T47
Download the bioisostere molecule30.sdf
Download the bioisostere molecule9.sdf
Link to the original publication
Download Molecule MRTX1133: 7t47_B_6IC.sdf extracted from the PDB complex 7T47
Download the bioisostere molecule30.sdf
Download the bioisostere molecule9.sdf
Link to the original publication
Inputs:Reference 7t47_B_6IC.sdf
Fragment to replace is number 3 in make_fgts_aggreg.sdf
Database pdb2
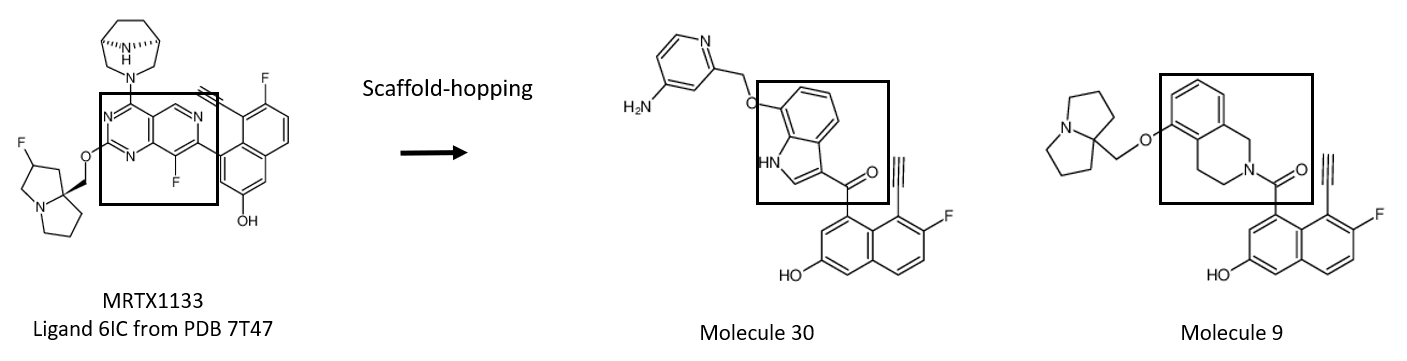
ResultsIn this tutorial, results show that the molecule number 100 in ordered-bioisosteres-pdb2.sdf corresponds to a very similar structure of the molecule 30 in the publication
Download best aligned bioisosteres ordered-bioisosteres-pdb2.sdf (399) and scores ordered-bioisosteres-pdb2-score.txt (more information on scores).To visualize superimposition results: 3Dviewer
You can install and use DataWarrior (free software) to read the SDF file content as a table (available for Windows/Linux/MacOS-X)Note: it has been observed that the minimization of some molecular structures is not sufficient with the standard process (RDKit, MMFF94 force field, 2000 Minimization cycles). This can sometimes cause some structure distortion).
Run completed
 |
Copyright © 2019 Université Côte d'Azur CNRS - All rights reserved
| Contact | |
 |