| | ||||||||||
| 1 | INN | Class | Route (list) | PK parameters= Cmax; Tmax; F: bioavailability; t1/2: half-life; VD: volume of distribution; Cl: clearance; PPB: plasma protein binding;(EQN means that value was calculated using VD=(Cl*t1/2)/0.693) | Primary Target and PDB code of Protein-Drug complex | Targets from DrugCentral | Links | |||
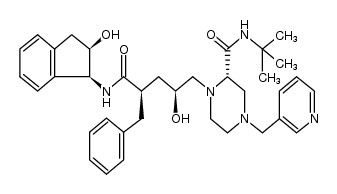
| INDINAVIR DISCONTINUED
| INDINAVIR | ANTIINFECTIVES | ORAL | Cmax 12 NANOMOLAR (800 MILLIGRAM EVERY 8 HOURS) VD 53.3 LITER (65 KILOGRAM) PPB 60 PERCENT Cl 70.2 LITER / HOUR (65 KILOGRAM) HT 1.8 HOUR SOLUBILITY GOOD IN WATER | HIV-1 PROTEASE PDB 1SGU (COMPARING THE ACCUMULATION OF ACTIVE SITE AND NON-ACTIVE SITE MUTATIONS IN THE HIV-1 PROTEASE) LIGAND CODE = MK1 (link to the list of PDB complexes) Download experimental 3D coordinates of MK1 with added hydrogens | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE INHIBITOR CHEMBL243 HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE Q72874 POL POLYPROTEIN HUMAN IMMUNODEFICIENCY VIRUS 1 ENZYME PROTEASE ASPARTIC AA A2A | EMA | |||